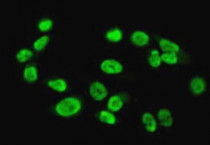
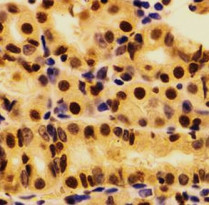
ARG57845
anti-Ku80 antibody
anti-Ku80 antibody for ICC/IF,IHC-Formalin-fixed paraffin-embedded sections,Immunoprecipitation,Western blot and Human,Mouse,Rat
Overview
| Product Description | Rabbit Polyclonal antibody recognizes Ku80 |
|---|---|
| Tested Reactivity | Hu, Ms, Rat |
| Tested Application | ICC/IF, IHC-P, IP, WB |
| Host | Rabbit |
| Clonality | Polyclonal |
| Isotype | IgG |
| Target Name | Ku80 |
| Antigen Species | Human |
| Immunogen | Recombinant protein of Human Ku80. |
| Conjugation | Un-conjugated |
| Alternate Names | double-strand-break rejoining; Thyroid-lupus autoantigen; Nuclear factor IV; KU80; DNA repair protein XRCC5; KARP1; Lupus Ku autoantigen protein p86; EC 3.6.4.-; CTCBF; CTC85; ATP-dependent DNA helicase 2 subunit 2; X-ray repair cross-complementing protein 5; ATP-dependent DNA helicase II 80 kDa subunit; CTC box-binding factor 85 kDa subunit; KARP-1; X-ray repair complementing defective repair in Chinese hamster cells 5; 86 kDa subunit of Ku antigen; KUB2; NFIV; TLAA; Ku80; Ku86 |
Application Instructions
| Application Suggestion |
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Application Note | * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. |
Properties
| Form | Liquid |
|---|---|
| Purification | Affinity purified. |
| Buffer | PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. |
| Preservative | 0.02% Sodium azide |
| Stabilizer | 50% Glycerol |
| Storage Instruction | For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. |
| Note | For laboratory research only, not for drug, diagnostic or other use. |
Bioinformation
| Database Links |
Swiss-port # P13010 Human X-ray repair cross-complementing protein 5 Swiss-port # P27641 Mouse X-ray repair cross-complementing protein 5 |
|---|---|
| Gene Symbol | XRCC5 |
| Gene Full Name | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| Background | The protein encoded by this gene is the 80-kilodalton subunit of the Ku heterodimer protein which is also known as ATP-dependant DNA helicase II or DNA repair protein XRCC5. Ku is the DNA-binding component of the DNA-dependent protein kinase, and it functions together with the DNA ligase IV-XRCC4 complex in the repair of DNA double-strand break by non-homologous end joining and the completion of V(D)J recombination events. This gene functionally complements Chinese hamster xrs-6, a mutant defective in DNA double-strand break repair and in ability to undergo V(D)J recombination. A rare microsatellite polymorphism in this gene is associated with cancer in patients of varying radiosensitivity. [provided by RefSeq, Jul 2008] |
| Function | Single-stranded DNA-dependent ATP-dependent helicase. Has a role in chromosome translocation. The DNA helicase II complex binds preferentially to fork-like ends of double-stranded DNA in a cell cycle-dependent manner. It works in the 3'-5' direction. Binding to DNA may be mediated by XRCC6. Involved in DNA non-homologous end joining (NHEJ) required for double-strand break repair and V(D)J recombination. The XRCC5/6 dimer acts as regulatory subunit of the DNA-dependent protein kinase complex DNA-PK by increasing the affinity of the catalytic subunit PRKDC to DNA by 100-fold. The XRCC5/6 dimer is probably involved in stabilizing broken DNA ends and bringing them together. The assembly of the DNA-PK complex to DNA ends is required for the NHEJ ligation step. In association with NAA15, the XRCC5/6 dimer binds to the osteocalcin promoter and activates osteocalcin expression. The XRCC5/6 dimer probably also acts as a 5'-deoxyribose-5-phosphate lyase (5'-dRP lyase), by catalyzing the beta-elimination of the 5' deoxyribose-5-phosphate at an abasic site near double-strand breaks. XRCC5 probably acts as the catalytic subunit of 5'-dRP activity, and allows to 'clean' the termini of abasic sites, a class of nucleotide damage commonly associated with strand breaks, before such broken ends can be joined. The XRCC5/6 dimer together with APEX1 acts as a negative regulator of transcription. [UniProt] |
| Cellular Localization | Chromosome, Nucleus, nucleolus. [UniProt] |
| Calculated MW | 83 kDa |
| PTM | Phosphorylated on serine residues. Phosphorylation by PRKDC may enhance helicase activity. Sumoylated. Ubiquitinated by RNF8 via 'Lys-48'-linked ubiquitination following DNA damage, leading to its degradation and removal from DNA damage sites (PubMed:22266820). Ubiquitinated by RNF138, leading to remove the Ku complex from DNA breaks (PubMed:26502055). [UniProt] |
Images (2) Click the Picture to Zoom In